In the world of digital PCR (dPCR), there are words that appear frequently in scientific publications. Knowing and understanding these common terms is essential in order to understand the topic of dPCR and to understand and contribute to the scientific literature. Discover the terminology of dPCR explained in simple terms to gain a better understanding of the novel quantification method.
Prereaction
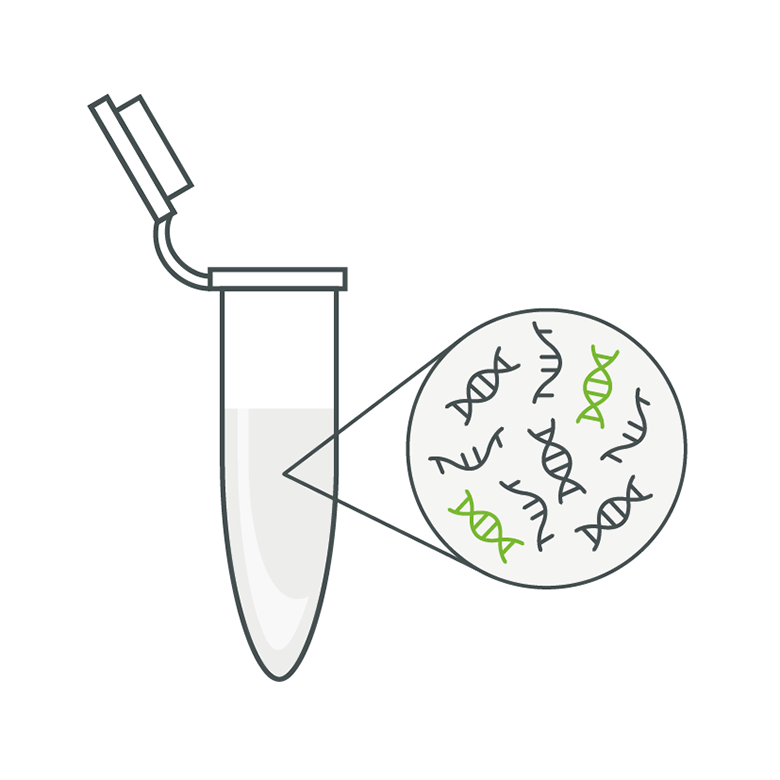
The main components of the PCR mix, which contains the initial volume that is prepared before splitting into small reactions. It contains all the necessary components such as master mix (polymerase, buffer, etc.), dNTPs (building blocks for synthesising new DNA strands), specific assay components (primers and probes) and the DNA template.
Partition (droplet; chamber)
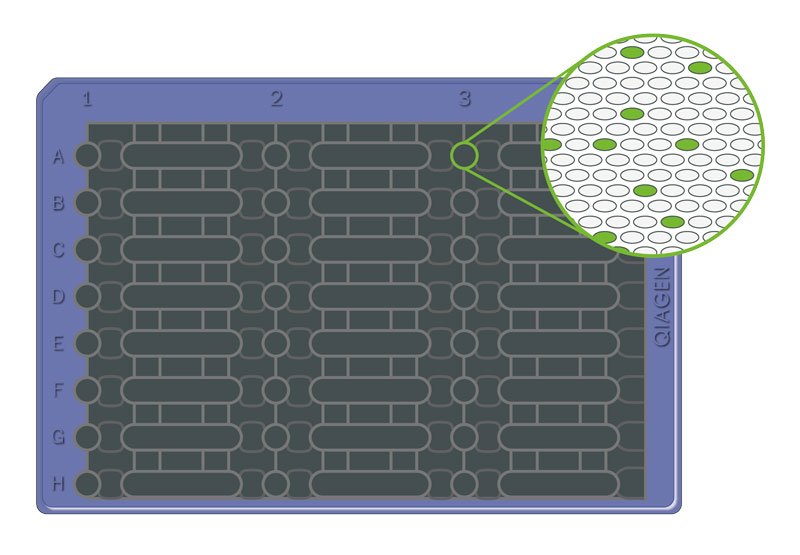
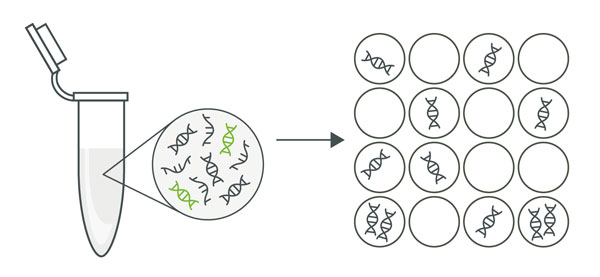
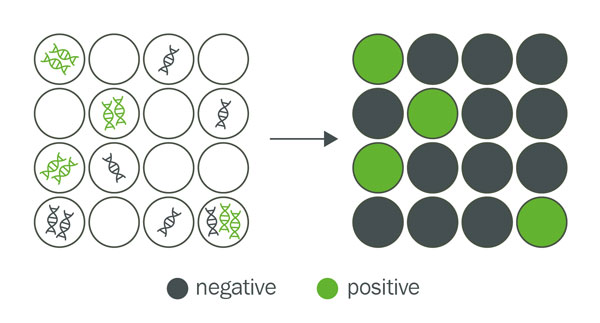
A partition is a single, small reaction unit that is created when the overall reaction is divided into many smaller ones. Each of these units can be either positive (with target DNA) or negative (without target DNA) after the reaction.
Reaction
The total volume of the partitions measured. It is calculated by multiplying the number of total partitions (accepted partitions; analysable partitioning) used for quantification by the volume of each partition. The volume of the partitions must be even (volume equality) to ensure accurate and reproducible results. The unit output by the software is copies per input volume.
Partitioning
The process of dividing a PCR reaction mixture into thousands of small, individual reactions (partitions), each acting as an independent PCR reaction. Each small reaction is like a mini-test to look for the target DNA. This can be achieved using droplets (droplet digital PCR, ddPCR) or chambers (microplate dPCR).
Absolute Quantification
A method that enables the direct counting of DNA or RNA molecules without reference or
comparison (standard curves). dPCR provides an accurate (absolute) count of initial target
molecules present and enables precise measurement of nucleic acid concentration.
Poisson (Poisson Distribution)
A statistical model used in dPCR to describe the random distribution of target molecules
across the partitions. When the DNA molecules are distributed across the partitions, each
partition receives either no molecule (negative partition) or one or more molecules
(positive partition). By applying the Poisson distribution, the original concentration of
the target molecules in the sample can be deduced from the number of positive and negative
partitions.
Positive/negative signal
Each partition in a dPCR experiment is classified as positive if it contains at least one or more target sequences and shows amplification, or as negative if it does not contain the target sequence and shows no amplification.
Dynamic Range
The range of target molecule concentrations that dPCR can accurately quantify. The dynamic
range reflects the ability to measure both high and low concentrations of nucleic acids with
high precision.
Copy Number Variation (CNV)
Differences in the number of copies of a particular gene or DNA segment in the genome. dPCR
is used to accurately quantify these differences (variations), which are important in many
genetic studies and disease diagnostics.
Multiplexing
The ability to simultaneously amplify and quantify multiple target DNA or RNA sequences in a
single dPCR reaction. This is achieved by using different sets of primers and probes, each
specific to a different target, and typically labeled with various fluorescent dyes. This
approach increases the efficiency and throughput of dPCR assays.
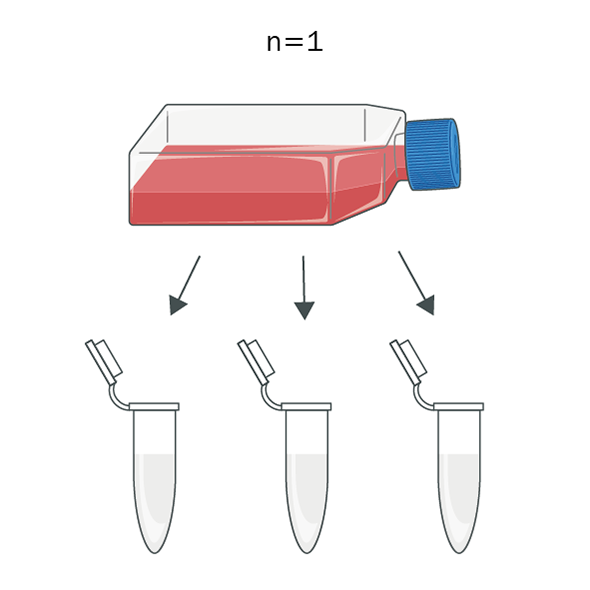
Technical Replicates
Multiple identical reactions conducted to assess the precision and reproducibility of dPCR results. They help in identifying and minimizing random errors. In other words, a test that is repeated with the same sample to ensure that the results are consistent and reliable.
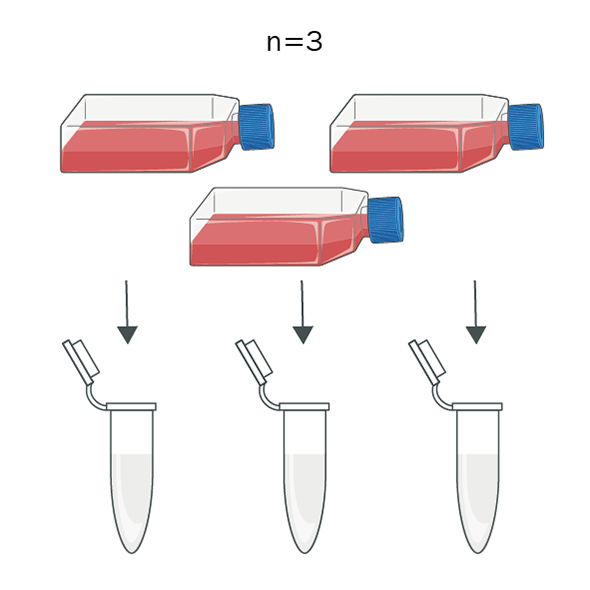
Biological Replicates
Independent samples from the same biological source are tested to assess biological variability and ensure that the results are representative and reproducible. This ensures that the test produces accurate and consistent results across different biological samples.
Dead Volume
The difference between the volume of the prereaction and the volume of the reaction. This
includes losses that occur during preparation and partitioning. It is the volume that is not
directly involved in the reaction but remains in the reaction chamber or dPCR plate and is
not measured.
Fluorescence Intensity (Fluorescence Amplitude, Endpoint
Fluorescence, Relative Fluorescence Unit)
The intensity of the fluorescence signal of a partition. This signal is used to determine
whether a partition is positive or negative.
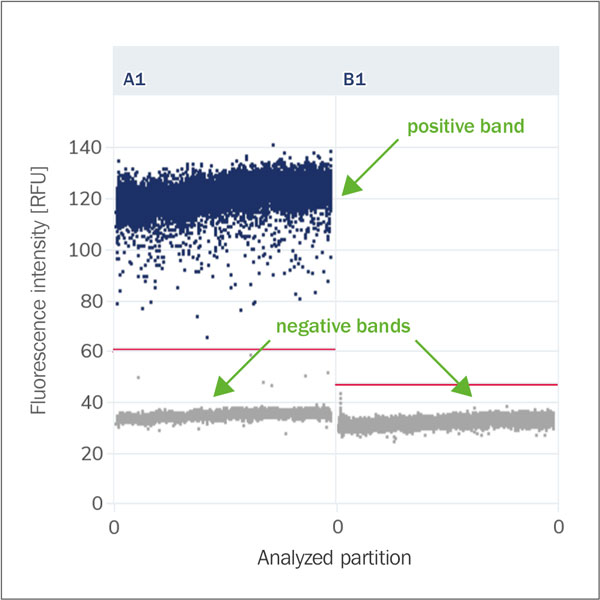
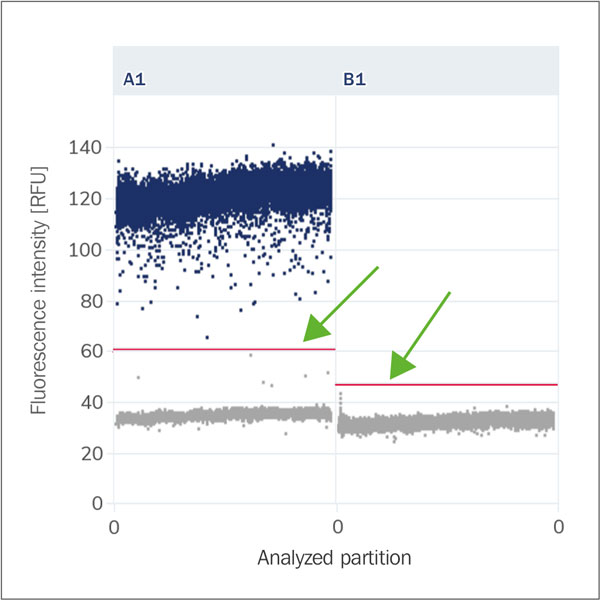
Positive and negative bands
The terms positive band and negative band refer to the areas in the scatter diagram that represent the fluorescence intensity of positive and negative partitions. The bands help to visualise and interpret the results of the dPCR. A clear separation between the positive and negative bands is crucial to ensure accurate and reliable results. A well-defined positive band means that the PCR was successful, while a well-defined negative band confirms that there is no cross-contamination or non-specific amplification.
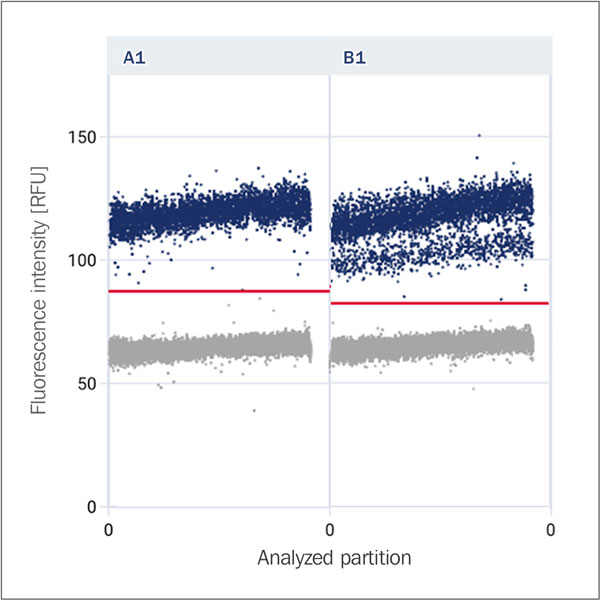
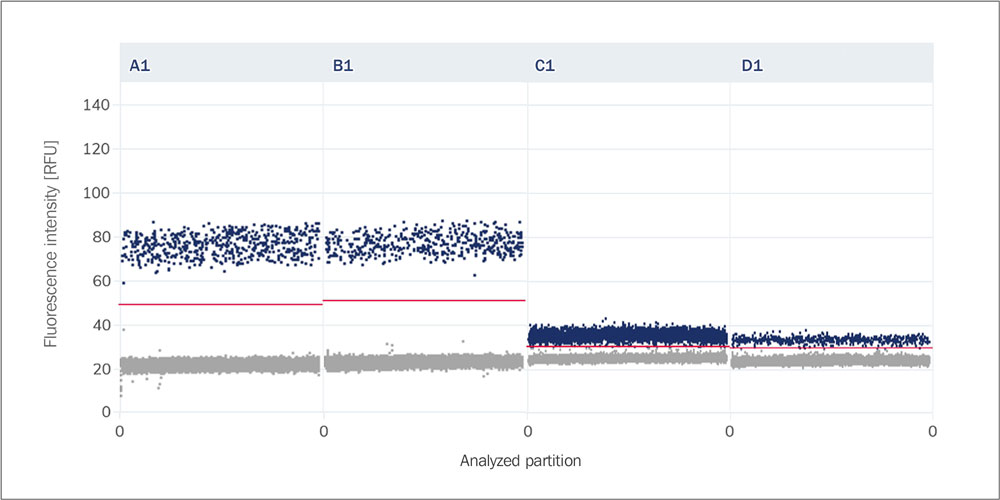
Double-banding
Double-banding describes the occurrence of two different fluorescence bands within the positive partitions in a dPCR reaction, which may indicate variability in amplification or the presence of different target molecules.
Threshold
The dividing line that separates the clusters of the partitions based on their fluorescence amplitude. This threshold value is crucial for accurately quantifying the number of target molecules in a sample. The automatic threshold refers to a software-determined cut-off value that distinguishes between positive and negative partitions based on their fluorescence intensity.
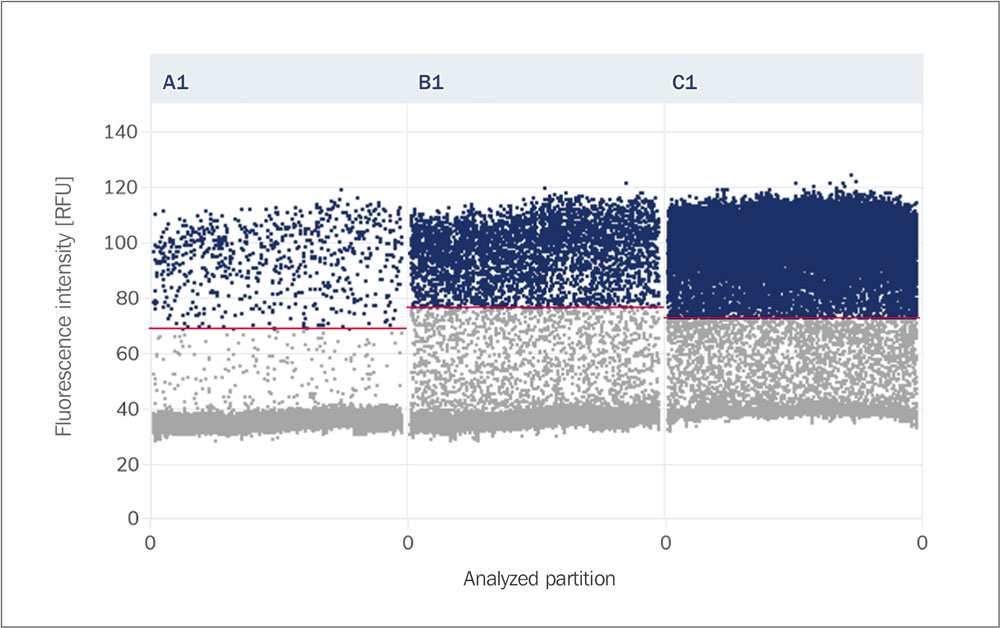
Rain
Partitions that lie in the area between the positive and negative clusters. These can represent uncertainties (no clear separation by fluorescence intensity) or transitions (overlapping of signals).
Baseline (Fluorescence Noise, Background)
The baseline fluorescence of negative partitions that do not contain target DNA. This serves
as a reference value. It represents the natural background or noise generated by the
reaction mixture, the reaction chambers, or the measurement system itself.
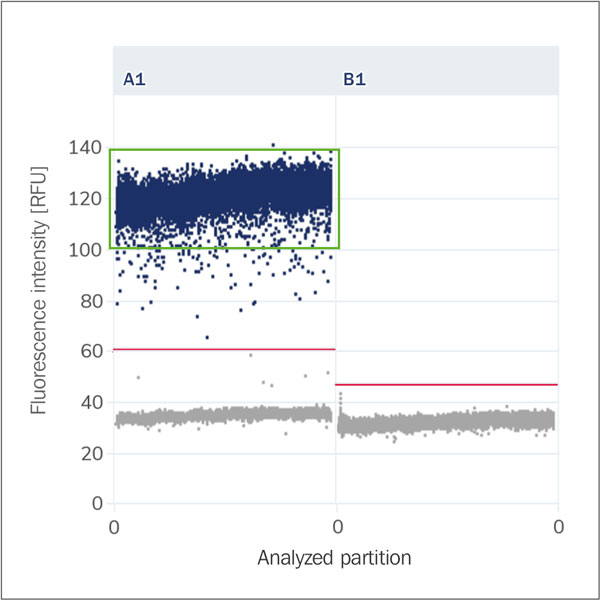
Cluster (Population, Droplet Population)
A group of partitions that lie within a similar range in the scatter plot based on their amplitude. Positive and negative partitions each form their own clusters.
Peak Resolution (Separability Score, Amplitude)
Peak resolution is a measure of how well positive and negative partitions are separated based
on their fluorescence intensity. Its purpose is to assess the clarity and distinctness of
the fluorescence signals, ensuring that positive and negative signals are easily
distinguishable. This metric plays a crucial role in evaluating the performance of the dPCR
assay. A high peak resolution indicates that the positive and negative clusters are well
separated, which leads to more reliable and accurate results.
Signal-to-noise ratio (SNR)
The signal-to-noise ratio measures how strong the desired signal (e.g. the fluorescence of amplified DNA molecules) is compared to the background noise (unwanted fluorescence signals that are not related to the target DNA). High peak resolution usually also requires a good signal-to-noise ratio, as a strong signal with little noise can lead to better separated peaks. However, in some cases, even with good SNR, the peaks may not be clearly separated, indicating other problems such as non-specific amplification or variability in the sample.
Swelling
An effect in which the partitions become uneven by varying the size of the individual
partitions or droplets, which can affect the accuracy of the quantification.
Standardization
Ensuring consistency and accuracy across different dPCR experiments and instruments.
Stay tuned and discover more articles about molecular biological detection methods and their applications in research, diagnostics and industry!
Be the first to know and subscribe to our newsletter below!
Sources:
dMIQE Group; Huggett JF. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin Chem. 2020 Aug 1;66(8):1012-1029. doi: 10.1093/clinchem/hvaa125. Erratum in: Clin Chem. 2020 Nov 1;66(11):1464. doi: 10.1093/clinchem/hvaa219. PMID: 3274645
Morley AA. Digital PCR: A brief history. Biomol Detect Quantif. 2014 Aug 15;1(1):1-2. doi: 10.1016/j.bdq.2014.06.001. PMID: 27920991; PMCID: PMC5129430